Publications
Publications I contributed to.
2025
-
 Global Context-aware Representation Learning for Spatially Resolved TranscriptomicsYunhak Oh, Junseok Lee, Yeongmin Kim, and 3 more authors2025
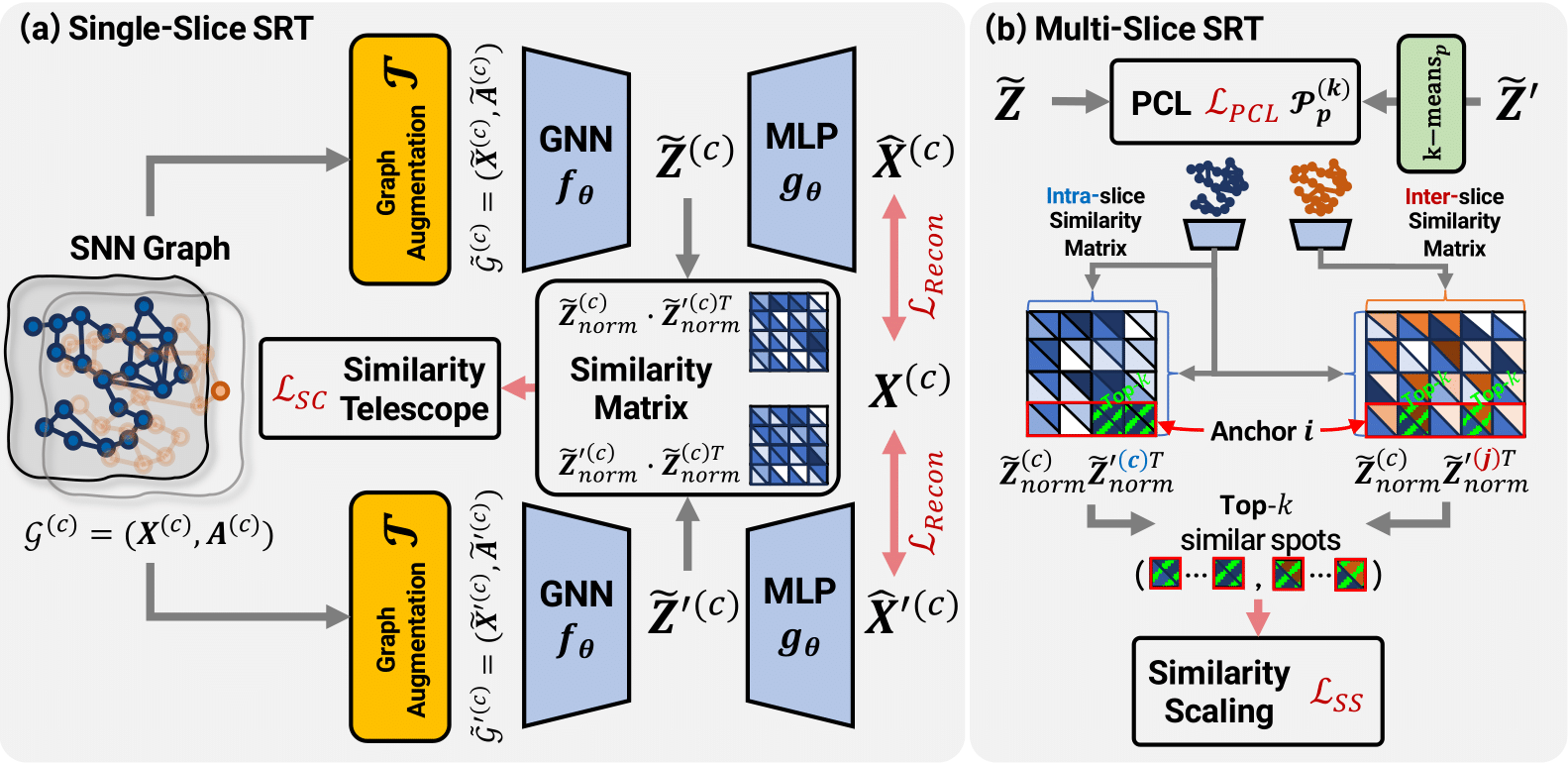
Global Context-aware Representation Learning for Spatially Resolved TranscriptomicsYunhak Oh, Junseok Lee, Yeongmin Kim, and 3 more authors2025Spatially Resolved Transcriptomics (SRT) is a cutting-edge technique that captures the spatial context of cells within tissues, enabling the study of complex biological networks. Recent graphbased methods leverage both gene expression and spatial information to identify relevant spatial domains. However, these approaches fall short in obtaining meaningful spot representations, especially for spots near spatial domain boundaries, as they heavily emphasize adjacent spots that have minimal feature differences from an anchor node. To address this, we propose Spotscape, a novel framework that introduces the Similarity Telescope module to capture global relationships between multiple spots. Additionally, we propose a similarity scaling strategy to regulate the distances between intra- and inter-slice spots, facilitating effective multi-slice integration. Extensive experiments demonstrate the superiority of Spotscape in various downstream tasks, including single-slice and multi-slice scenarios. Our code is available at the following link: https: //github.com/yunhak0/Spotscape.
2024
-
 Single-cell RNA sequencing data imputation using bi-level feature propagationJunseok Lee, Sukwon Yun, Yeongmin Kim, and 3 more authorsBriefings in Bioinformatics, 2024
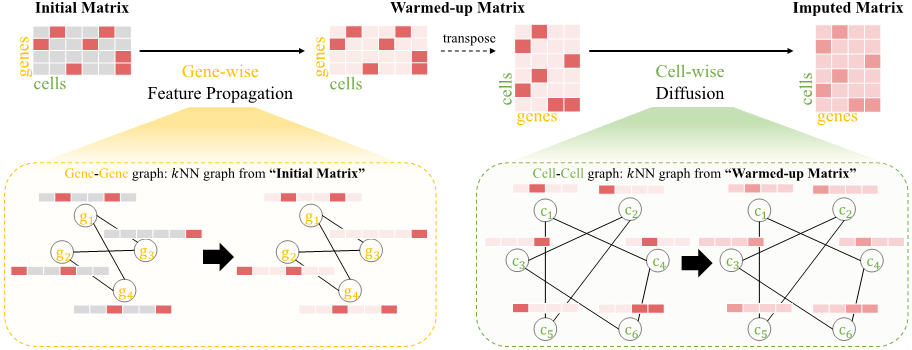
Single-cell RNA sequencing data imputation using bi-level feature propagationJunseok Lee, Sukwon Yun, Yeongmin Kim, and 3 more authorsBriefings in Bioinformatics, 2024Single-cell RNA sequencing (scRNA-seq) enables the exploration of cellular heterogeneity by analyzing gene expression profiles in complex tissues. However, scRNA-seq data often suffer from technical noise, dropout events and sparsity, hindering downstream analyses. Although existing works attempt to mitigate these issues by utilizing graph structures for data denoising, they involve the risk of propagating noise and fall short of fully leveraging the inherent data relationships, relying mainly on one of cell–cell or gene–gene associations and graphs constructed by initial noisy data. To this end, this study presents single-cell bilevel feature propagation (scBFP), two-step graph-based feature propagation method. It initially imputes zero values using non-zero values, ensuring that the imputation process does not affect the non-zero values due to dropout. Subsequently, it denoises the entire dataset by leveraging gene–gene and cell–cell relationships in the respective steps. Extensive experimental results on scRNA-seq data demonstrate the effectiveness of scBFP in various downstream tasks, uncovering valuable biological insights.
-
 A machine learning approach using conditional normalizing flow to address extreme class imbalance problems in personal health recordsYeongmin Kim, Wongyung Choi, Woojeong Choi, and 7 more authorsBioData Mining, 2024
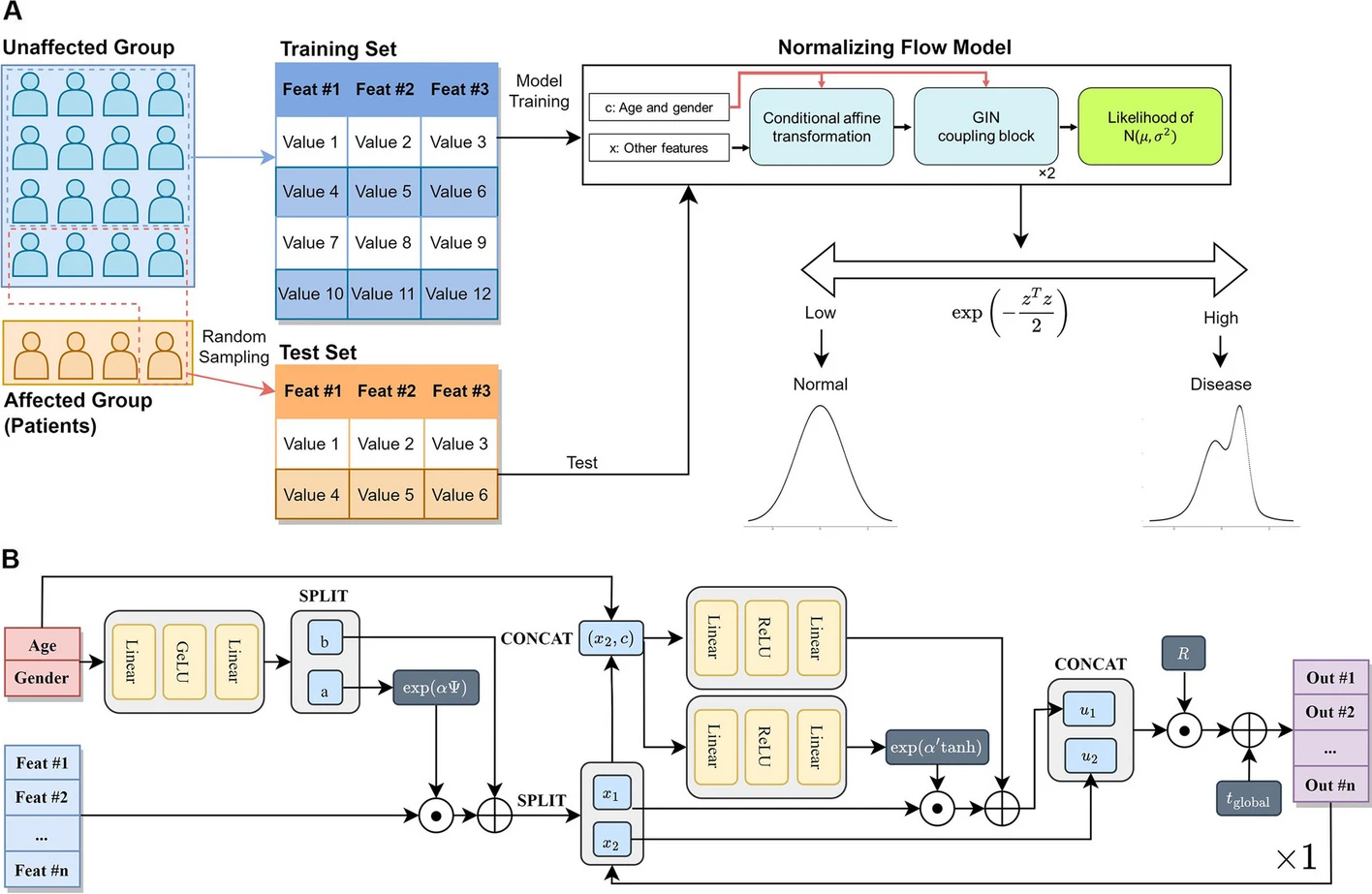
A machine learning approach using conditional normalizing flow to address extreme class imbalance problems in personal health recordsYeongmin Kim, Wongyung Choi, Woojeong Choi, and 7 more authorsBioData Mining, 2024Supervised machine learning models have been widely used to predict and get insight into diseases by classifying patients based on personal health records. However, a class imbalance is an obstacle that disrupts the training of the models. In this study, we aimed to address class imbalance with a conditional normalizing flow model, one of the deep-learning-based semi-supervised models for anomaly detection. It is the first introduction of the normalizing flow algorithm for tabular biomedical data. We collected personal health records from South Korean citizens (n = 706), featuring genetic data obtained from direct-to-customer service (microarray chip), medical health check-ups, and lifestyle log data. Based on the health check-up data, six chronic diseases were labeled (obesity, diabetes, hypertriglyceridemia, dyslipidemia, liver dysfunction, and hypertension). After preprocessing, supervised classification models and semi-supervised anomaly detection models, including conditional normalizing flow, were evaluated for the classification of diabetes, which had extreme target imbalance (about 2%), based on AUROC and AUPRC. In addition, we evaluated their performance under the assumption of insufficient collection for patients with other chronic diseases by undersampling disease-affected samples. While LightGBM (the best-performing model among supervised classification models) showed AUPRC 0.16 and AUROC 0.82, conditional normalizing flow achieved AUPRC 0.34 and AUROC 0.83 during fifty evaluations of the classification of diabetes, whose base rate was very low, at 0.02. Moreover, conditional normalizing flow performed better than the supervised model under a few disease-affected data numbers for the other five chronic diseases – obesity, hypertriglyceridemia, dyslipidemia, liver dysfunction, and hypertension. For example, while LightGBM performed AUPRC 0.20 and AUROC 0.75, conditional normalizing flow showed AUPRC 0.30 and AUROC 0.74 when predicting obesity, while undersampling disease-affected samples (positive undersampling) lowered the base rate to 0.02. Our research suggests the utility of conditional normalizing flow, particularly when the available cases are limited, for predicting chronic diseases using personal health records. This approach offers an effective solution to deal with sparse data and extreme class imbalances commonly encountered in the biomedical context.
2022
-
 Altub: Alternating training method to update base distribution of normalizing flow for anomaly detectionYeongmin Kim, Huiwon Jang, DongKeon Lee, and 1 more authorarXiv preprint arXiv:2210.14913, 2022
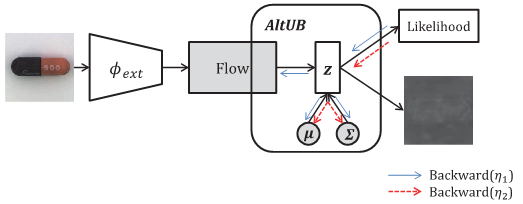
Altub: Alternating training method to update base distribution of normalizing flow for anomaly detectionYeongmin Kim, Huiwon Jang, DongKeon Lee, and 1 more authorarXiv preprint arXiv:2210.14913, 2022Unsupervised anomaly detection is coming into the spotlight these days in various practical domains due to the limited amount of anomaly data. One of the major approaches for it is a normalizing flow which pursues the invertible transformation of a complex distribution as images into an easy distribution as N(0, I). In fact, algorithms based on normalizing flow like FastFlow and CFLOW-AD establish state-of-the-art performance on unsupervised anomaly detection tasks. Nevertheless, we investigate these algorithms convert normal images into not N(0, I) as their destination, but an arbitrary normal distribution. Moreover, their performances are often unstable, which is highly critical for unsupervised tasks because data for validation are not provided. To break through these observations, we propose a simple solution AltUB which introduces alternating training to update the base distribution of normalizing flow for anomaly detection. AltUB effectively improves the stability of performance of normalizing flow. Furthermore, our method achieves the new state-of-the-art performance of the anomaly segmentation task on the MVTec AD dataset with 98.8% AUROC.


